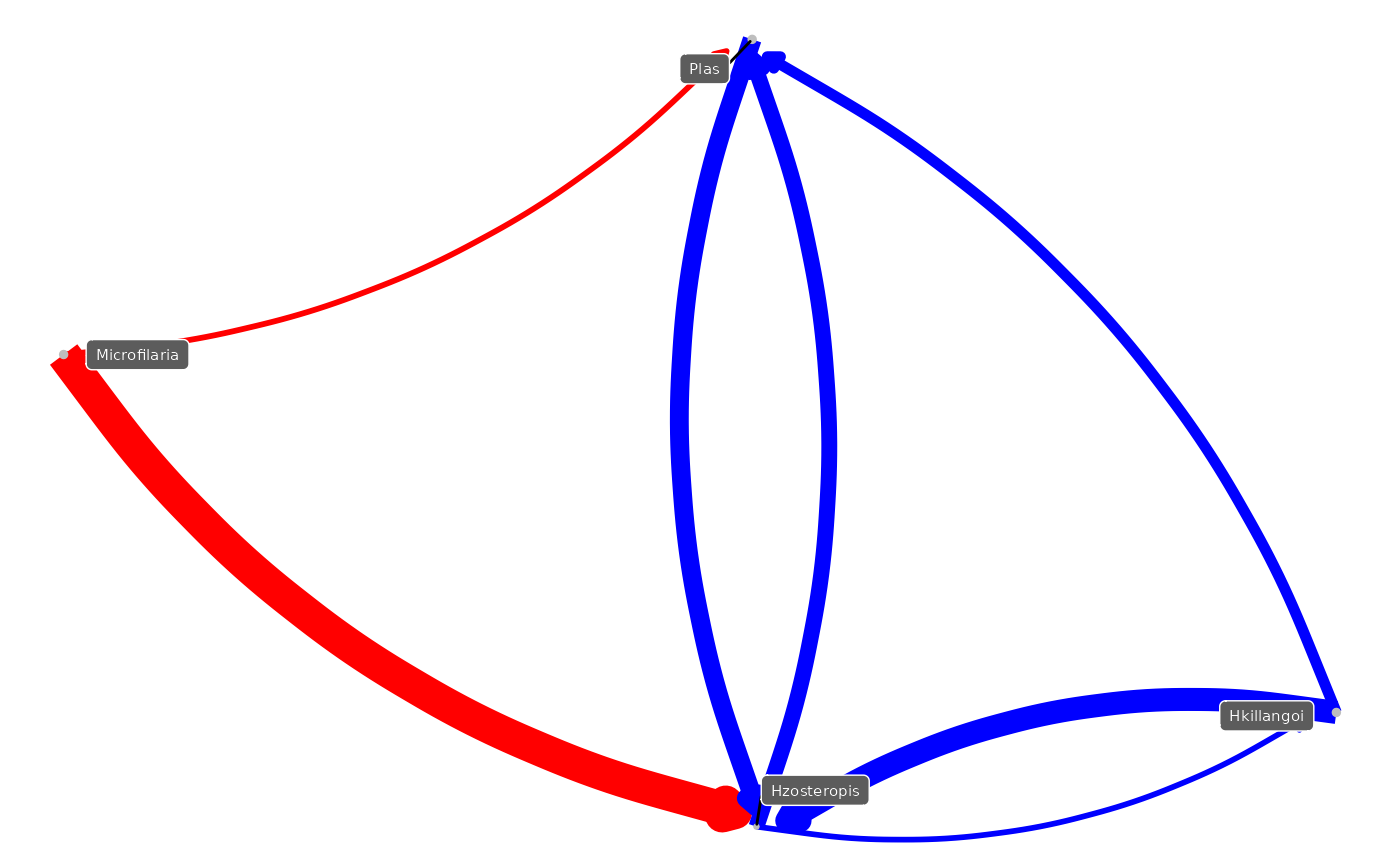
This function generates a co-occurrence network from a provided list and calculates strength and directionality of the relationships. The output can be passed to igraph to plot a directed acyclic graph (DAG).
Arguments
- mrBootstrap_obj
A list of bootstrapped partial dependencies output from
mrBootstrap().
Examples
library(tidymodels)
#> ── Attaching packages ────────────────────────────────────── tidymodels 1.4.1 ──
#> ✔ broom 1.0.10 ✔ recipes 1.3.1
#> ✔ dials 1.4.2 ✔ rsample 1.3.1
#> ✔ dplyr 1.1.4 ✔ tailor 0.1.0
#> ✔ ggplot2 4.0.0 ✔ tidyr 1.3.1
#> ✔ infer 1.0.9 ✔ tune 2.0.1
#> ✔ modeldata 1.5.1 ✔ workflows 1.3.0
#> ✔ parsnip 1.3.3 ✔ workflowsets 1.1.1
#> ✔ purrr 1.2.0 ✔ yardstick 1.3.2
#> ── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
#> ✖ purrr::discard() masks scales::discard()
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ✖ recipes::step() masks stats::step()
library(igraph)
#>
#> Attaching package: ‘igraph’
#> The following object is masked from ‘package:tidyr’:
#>
#> crossing
#> The following objects are masked from ‘package:purrr’:
#>
#> compose, simplify
#> The following objects are masked from ‘package:dplyr’:
#>
#> as_data_frame, groups, union
#> The following objects are masked from ‘package:dials’:
#>
#> degree, neighbors
#> The following objects are masked from ‘package:future’:
#>
#> %->%, %<-%
#> The following objects are masked from ‘package:stats’:
#>
#> decompose, spectrum
#> The following object is masked from ‘package:base’:
#>
#> union
library(ggnetwork)
mrIML_rf <- mrIML::mrIML_bird_parasites_RF
mrIML_rf_boot <- mrIML_rf %>%
mrBootstrap()
#>
|
| | 0%
|
|== | 2%
|
|==== | 5%
|
|===== | 8%
|
|======= | 10%
|
|========= | 12%
|
|========== | 15%
|
|============ | 18%
|
|============== | 20%
|
|================ | 22%
|
|================== | 25%
|
|=================== | 28%
|
|===================== | 30%
|
|======================= | 32%
|
|======================== | 35%
|
|========================== | 38%
|
|============================ | 40%
|
|============================== | 42%
|
|================================ | 45%
|
|================================= | 48%
|
|=================================== | 50%
|
|===================================== | 52%
|
|====================================== | 55%
|
|======================================== | 58%
|
|========================================== | 60%
|
|============================================ | 62%
|
|============================================== | 65%
|
|=============================================== | 68%
|
|================================================= | 70%
|
|=================================================== | 72%
|
|==================================================== | 75%
|
|====================================================== | 78%
|
|======================================================== | 80%
|
|========================================================== | 82%
|
|============================================================ | 85%
|
|============================================================= | 88%
|
|=============================================================== | 90%
|
|================================================================= | 92%
|
|================================================================== | 95%
|
|==================================================================== | 98%
|
|======================================================================| 100%
assoc_net_filtered <- mrIML_rf_boot %>%
mrCoOccurNet() %>%
filter(mean_strength > 0.05)
# Convert to igraph
g <- graph_from_data_frame(
assoc_net_filtered,
directed = TRUE,
vertices = names(mrIML_rf$Data$Y)
)
E(g)$Value <- assoc_net_filtered$mean_strength
E(g)$Color <- ifelse(
assoc_net_filtered$direction == "negative",
"blue", "red"
)
# Convert the igraph object to a ggplot object with NMDS layout
gg <- ggnetwork(g)
# Plot the graph
ggplot(
gg,
aes(x = x, y = y, xend = xend, yend = yend)
) +
geom_edges(
aes(color = Color, linewidth = Value),
curvature = 0.2,
arrow = arrow(length = unit(5, "pt"), type = "closed")
) +
geom_nodes(
color = "gray",
size = degree(g, mode = "out") / 2
) +
scale_color_identity() +
theme_void() +
theme(legend.position = "none") +
geom_nodelabel_repel(
aes(label = name),
box.padding = unit(0.5, "lines"),
size = 2,
segment.colour = "black",
colour = "white",
fill = "grey36"
)